Material on this page is also available as a recorded webinar.
See also About the Yosemite Project
Contents
Introduction
Today’s healthcare information systems are a Tower of Babel, with a wide variety of data formats, data models and vocabularies that do not interoperate well. This lack of interoperability impacts all aspects of healthcare, from patient care to research to administration, and wastes an estimated $30 billion USD per year.
Imagine what we could do in a world where all healthcare systems spoke the same language, with the same meanings covering all of healthcare. That’s known as semantic interoperability, or the ability of computer systems to exchange data with unambiguous shared meaning.
The mission of the Yosemite Project is to achieve semantic interoperability of all structured healthcare information, using RDF as a universal information representation.
Aside: Unstructured healthcare information, such as a doctor’s dictated note, is also very important, but it presents a different challenge due to the inherent ambiguity of human language to a computer. However, as Natural Language Processing (NLP) technology improves, more unstructured data will be interpretable as structured data via NLP.
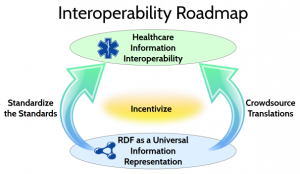
To reach this ambitious goal, the Yosemite Project articulates a technical roadmap with two major themes: Standardize the Standards and Crowdsource Translations. The roadmap is based on the use of RDF as a universal information representation.
RDF as a Universal Information Representation
Resource Description Framework (RDF) is an information representation language based on formal logic. An international standard developed by the World Wide Web Consortium (W3C). It allows information to be written in various forms and captured as information content — logical assertions — independent of data format. RDF has been used for over 10 years in a wide variety of domains, ranging from medical research to military intelligence and many others.
Note for techies: The term RDF is used herein to encompass the family of W3C standards that are typically used together when working with RDF — including RDF, RDFS and OWL.
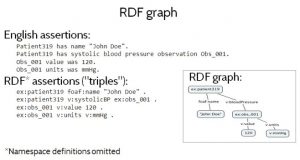
Figure 1 shows a small RDF graph — a set of RDF data — both as a diagram and as a set of assertions or “triples” written in Turtle format. Turtle namespaces have been omitted for brevity. Figure 1 (top) also shows an informal interpretation, in English prose, of the RDF assertions in the graph. Informally, the RDF triples say that some Patient319 has a name John Doe and a systolic blood pressure observation Obs_001, which recorded a pressure of value of 120 millimeters of mercury.
One important feature of RDF is that it captures information content independent of syntax or data formats.
Several different data formats are available for writing RDF, including Turtle, N-Triples, JSON-LD (a JSON-based syntax) and RDF/XML (an XML-based syntax). This means that the exact same information content be written using any of these RDF formats. But conversely, it also means that the information content from any data format — not only RDF data formats — can be represented in RDF, by providing a mapping from that non-RDF data format to any of the standard RDF formats.
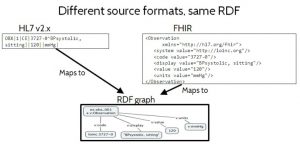
Figure 2 shows how the same information expressed in different non-RDF data formats can represent the same RDF information content, in spite of the fact that they are not standard RDF formats. On the left is some data formatted using HL7 v2.x, a widely used legacy standard that uses a delimited text string format. On the right it shows the equivalent data, formatted using FHIR XML, an emerging standard based on XML syntax. Neither of these are standard RDF data formats. But if a mapping is provided from these formats to a standard RDF format, then the RDF information content of the data can be determined. In Figure 2 they map to the exact same RDF information content, in spite of the fact that they originated in completely different non-RDF data formats. The ability to expose the intended information content, independent of data format, is important because it puts the emphasis on the meaning of the data — where it should be — rather than on insignificant syntactic issues.
This ability to capture information content, distinct from data format, is what enables RDF to act as a universal information representation. It also means that we don’t have to discard our existing data formats. We can still use them, but each one can have an RDF equivalent that unambiguously captures its intended information content. Furthermore, there is no need to explicitly exchange an RDF format over the wire, as long as the format that is exchanged has a standard RDF equivalent available, so that its information content can be understood in a universal way.
This ability to capture information content, distinct from data format, is what enables RDF to act as a universal information representation. It also means that we don’t have to discard our existing data formats. We can still use them, but each one can have an RDF equivalent that unambiguously captures its intended information content. Furthermore, there is no need to explicitly exchange an RDF format over the wire, as long as the format that is exchanged has a standard RDF equivalent available, so that its information content can be understood in a universal way.
For more details explaining why over 100 thought leaders in healthcare and technology believe that RDF is the “best available candidate” for this purpose, see also:
Standardize the Standards
The Unified Medical Language System (UMLS), created by the National Library of Medicine, currently catalogs over 100 standard vocabularies. To help address this bewildering diversity, the Office of National Coordinator (ONC) has published an interoperability standards advisory recommending a subset of about 30 of those vocabularies for use in the most common kinds of healthcare data exchanged in the USA. The advisory also includes clarifications to those standards to address cases where the standards are not specific enough to enable real interoperability. Even with this reduction, we still have a patchwork of partially overlapping and inconsistent standards that use different data formats, different data models, different vocabularies, and are not defined in a common, uniform and computable form.
One might think that the solution to this problem is to come up with a new standard to rule them all. But the naivete of that approach is nicely captured in an XKCD cartoon shown in Figure 3, How Standards Proliferate. Rather than truly solving the problem, a new standard typically ends up being yet another island, having its own conventions and its own sweet spot of applicability and use cases, with a significant amount of duplication and inconsistency between it and previous standards.
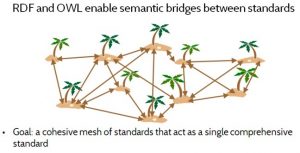
For this reason, the goal of the Yosemite Project and the use of RDF is not to eliminate or replace standards. Rather, the goal is to achieve a cohesive mesh of standards that can be used together, as though they constitute a single comprehensive standard. To do this, we need to standardize the standards.
The notion of standardizing the standards is to use RDF to achieve two goals. The first is to express computable semantic bridges between standards, by defining precise semantic relationships between standards. For example, two drug vocabularies might both include equivalent concepts of aspirin. Or one may define a concept that is broader or narrower than another, such as aspirin-in-general versus aspirin-tablets-325-mg. However, sometimes two concepts overlap without either one being strictly broader than the other. In such a case the concepts can be subdivided into smaller concepts, so that both of the original concepts can be expressed in terms of these common smaller concepts. The second goal in standardizing the standards is to facilitate convergence on common definitions across standards, by expressing those definitions in RDF, so that different standards can be used together harmoniously. By explicitly exposing the correspondences and differences between concepts in different standards, standards committees can more easily reference concepts from other standards, or adjust their own concepts to align.
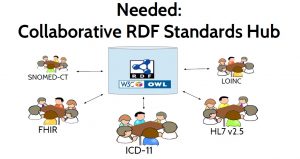
The purpose of a collaborative standards hub, depicted in Figure 5, is to facilitate and
accelerate the process of standards convergence. It would be used by those creating, managing, and revising the various healthcare information standards, and by implementers who need to use those standards. It would hold RDF definitions of the data models, vocabularies, and terminologies defined by those standards and it would expose their similarities and their differences in RDF to enable computable linkages between them. This hub could directly store the master version of a standard, but it should also be able to reference standards definitions that are defined elsewhere. It could provide metrics about these standards, and suggest related concepts to assist in linking between standards. It could also run reasoners to automatically check for logical inconsistencies both within a standard and across standards, and notify standards developers when an inconsistency is introduced. This collaborative standards hub might resemble a cross between GitHub, WikiData, CIMI Repository, UMLS, NCBO BioPortal and Web Protégé.
A collaborative standards hub should be easily accessible by providing a browser-based user interface. But it should also provide a RESTful API to facilitate integration with other tools and allow different groups to use other user interfaces, while still gaining the back-end benefits of using the standards hub. By keeping RDF “under the hood” — exposed to users only if they wish access to it — specialized front ends could be provided for use by medical domain experts with no knowledge of RDF.
One tool that takes an important step toward this vision of a collaborative standards hub is iCat — a custom front end that was used to define the ICD-11 standard. See Figure 6, ‘Web Protege tool for ICD-11’. Although iCat is based on Web Protégé and uses RDF internally, the user interface was specifically designed to be used by medical domain experts without knowledge of RDF. iCat has been used by over 270 medical experts around the world to define over 45,000 concepts with 17,000 links to external terminologies. Over 260,000 changes have been made through iCat in the course of developing ICD-11. However, iCat currently supports only ICD-11, rather than acting as a hub for multiple standards.
Healthcare is not the only industry facing need for information interoperability.
A parallel effort called FIBO (Financial Industry Business Ontology) has been undertaken in the financial industry to standardize the terms and definitions used for processing, modeling and reporting, based on RDF. The FIBO effort similarly seeks to define standard computable ontologies, and it has similarly recognized the need for a hub or central repository for FIBO ontologies, to facilitate collaboration and consistency. It would be nice if the same infrastructure could support healthcare, financial and other standards collaboration.
Another way that RDF facilitates standardizing the standards is that it helps avoid the bike shed effect — Parkinson’s Law of Triviality — because RDF does not care about data format, syntax, or even the names of things. It just cares about the information content and the relationships between concepts. This means that standards groups can avoid wasting so much time on things like syntax and names — decisions that are largely irrelevant to a computer — and focus on the meaning of the information content, which is where the focus should be.
Limitations of Standardization- Why Translation Is Also Needed
There are fundamentally two ways to achieve semantic interoperability: standardization or translation. With the standardization approach, interoperability is achieved by requiring all participants to speak the same language. In terms of computer systems, this means exchanging information using the same the data models and vocabularies — standardizing them. In contrast, with the translation approach, interoperability is achieved by translating between languages. In terms of computer systems, this means translating between data models and vocabularies.
Obviously we would prefer to avoid the overhead of translation and take the more efficient approach: standardization. Indeed, standardization is always preferable to the extent that it is feasible. However there are some fundamental limitations to the interoperability can be achieved using a standardization strategy alone.
One limitation is that the standards development process itself takes time. The more comprehensive a standard aims to be, the longer it takes to develop. There is an inherent trade-off between comprehensiveness, quality and timeliness: a standard can be developed quickly, at the expense of comprehensiveness, quality or both. Or a standard can be comprehensive, in which case it will either be poor quality or it will take forever to develop. Or a standard can be high quality, but the cost will be comprehensiveness, timeliness or both. This is the standards trilemma: it is not possible to create a standard that is simultaneously comprehensive, timely and high quality.
A second fundamental limitation of standardization is that modernization of computer systems to support a new standard requires substantial time and effort. Even if an ideal comprehensive standard were magically developed today, it would take years for existing systems to be updated. And since they cannot all be updated at the same time, different standards must be accommodated in the meantime — some systems running the new standard, while some run old standards.
A third significant limitation of standardization alone is that healthcare involves a wide variety of diverse use cases, requiring not only different kinds of data, but different data representations and data granularity. One size does not fit all.
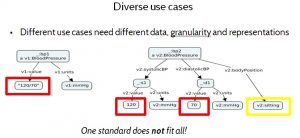
For example, Figure 9 Diverse Use Cases illustrates how a blood pressure data model may vary for different purposes. In the data model on the left, the systolic and diastolic values of the blood pressure measurement are represented as a single string, “120/70”, whereas the data model on the right represents them as two separate numbers — 120 and 70 — and it indicates the patient’s body position when the measurement was taken.
While the composite representation on the left is convenient for a doctor who is familiar with that format, the representation on the right is more convenient for a computer system that tracks blood pressure changes, because it does not need to parse the data apart into its constituent values. Furthermore the representation on the right has finer granularity — capturing body position — which may matter to some use cases, because it affects the interpretation of the data. A single standard cannot address all needs ideally.
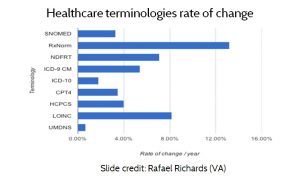
A fourth limitation of the standardization approach is that standards change. As AMIA president Dr. Douglas Fridsma has observed, “the only standards that do not change are the ones that nobody uses”. Dr. Rafael Richards at the VA examined the rate of change for several of the most common standard terminologies, (See Figure 10, Healthcare terminologies rate of change.) According to data published by the standards organizations themselves, the standards typically change from 4% to 8% per year. Again, since systems cannot all be updated at once, an effective interoperability strategy must accommodate multiple standards and versions.
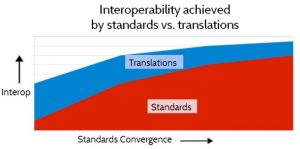
Because of these fundamental factors, a realistic strategy for semantic interoperability must address both standardization and translation. As standards become more standardized the relative proportion of interoperability that is achieved through standardization will increase, and the proportion achieved through translation will decrease, as depicted in Figure 11. Interoperability achieved by Standardization vs. Translation. But the need for translation will never be reduced to zero, for the fundamental reasons discussed above.
Crowdsource Translations
RDF can help with translation in a number of ways. One is that it supports inference, which is the process of deriving new facts from a given set of facts plus a set of inference rules. In RDF, this means using RDF inference rules to derive new RDF assertions from existing RDF assertions. Because inference takes one set of RDF assertions and produces a new set of RDF assertions, it can also be viewed as a form of data translation: the old RDF assertions are effectively translated into the new RDF assertions by the inference rules. For this reason, we also call them translation rules. This technique is often used when translating RDF data from one data model to another.
Another way that RDF can help with translation is by enabling translation rules to be more reusable. By operating on the same universal information representation — RDF — translation rules can be more easily shared, and mixed and matched.
For example, a source system that produces data in one format and data model — such as HL7 v2.x for example — may need to provide data to a target system that only understands a different format and data model, such as FHIR. This requires translating the data from the data format, data model and vocabulary of the source system, into the data format, data model and vocabulary of the target system.
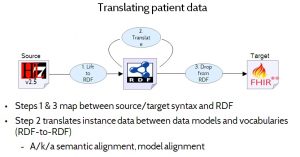
Figure 12 (‘Translating patient data’) illustrates how RDF’s ability to act as a universal information representation helps to solve this translation problem. The process involves three main steps:
- Step 1: Lift the data from the source format to RDF. This is a relatively simple, direct transliteration from one format to another, which does not attempt to transform or align data models or vocabularies. The resulting RDF will look very much like the original source data — having the same overall structure, and using the same vocabularies — but the syntax will be different.
- Step 2: Align the RDF data to the target data model and vocabulary. This is typically the hardest part. It involves translating from one form of RDF to another form of RDF, and it may actually involve multiple translation steps (signified by the loop in Figure 12). This step does whatever data model and vocabulary translation is needed, to go from the source data model and vocabulary to the target data model and vocabulary (still represented in RDF), potentially via a common intermediate data model and vocabulary.
- Step 3: Drop the data from RDF to the target format. This is the inverse of step 1. It performs a direct transliteration from RDF to the target format.
The terms lift and drop refer to the fact that RDF is considered a more abstract, higher level representation.
Three benefits are gained by factoring out the data format translations (steps 1 and 3) from the data model and vocabulary translations in step 2. One is that the model and vocabulary translation step can be simpler. Divide and conquer! Another is that diverse translators, written in different languages, can be used together, because they all act on RDF. A third is that these translators become more sharable and reusable, again because they all operate on RDF as a universal information representation.
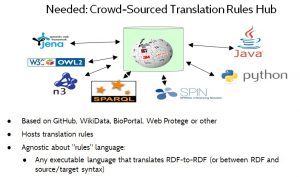
Figure 13 illustrates the idea of a crowd-sourced translation rules hub. This would be a web-based repository that allows translation rules — a/k/a translators or mapping rules — to be shared and reused.
Note for techies: The term translation rule herein refers to any executable artifact that performs data format, data model or vocabulary translation of instance data. Translation rules may be written either in a declarative or a procedural style, and may range from simple mapping tables, to path expressions, to inference rules, to executable code written in a general-purpose programming language.
The translation rules hub should be agnostic about the language that is used for expressing translation rules, to let the market decide which languages are preferred. For example, translation rules might be written in ShEx, SPARQL, OWL or even Python or JavaScript.
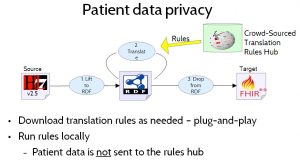
To ensure patient data privacy, patient data would not be uploaded to the translation rules hub. Instead, healthcare providers needing to perform data translation would download the translation rules and run them locally, as shown in Figure 14. Patient Data Privacy.
Incentivize
The final component of the Yosemite Roadmap is to incentivize interoperability. While this is not the main focus of the Yosemite Project — the main focus is the technical challenge of achieving interoperability — the need to incentivize interoperability is included it the roadmap because widespread interoperability cannot be achieved without it. The reason for this is that there is no natural business incentive for healthcare providers to share their data with their competitors (although they all want their competitors to share their data). While individual healthcare workers generally try to serve their patients’ best interests, healthcare providers as organizations operate are run as competitive businesses and set their own internal policies that their employees must follow. For this reason, carrot or stick policies are essential to ensure that healthcare providers will make their data interoperable. Policy makers must address this.
What will it cost to achieve semantic interoperability in healthcare? That is hard to estimate, and estimates will depend on what is included. Rough guesses might be in the range of $400 million per year for standardization and $300 million per year for translation, or $700 million per year total. As illustrated in Figure 15, that may sound expensive, but it is paltry when compared to the estimated $30000 million that is currently wasted each year because healthcare data is not yet interoperable.
Conclusion
The interoperability roadmap articulated by Yosemite Project is ambitious but feasible. It describes a technical and social process, based on standardizing the standards and crowdsourcing translations, that enables the full diversity of structured healthcare information to become interoperable. Based on RDF as a universal information representation, it allows the intended information content of any structured healthcare data to be exposed in a uniform way. This facilitates the convergence of healthcare standards around a common set of concepts that span all healthcare standards, and it facilitates the translation of healthcare data from one form to another when necessary. Although the effort needed to achieve interoperability would be considerable, the savings both in money and improved healthcare would far exceed the cost.